wwPDB 2023 News
Contents
12/18/2023
Improved Depositor Experience Using ORCiD
 ORCiD login button at in OneDep circled in red
ORCiD login button at in OneDep circled in redSince March 2023, depositors can use their ORCiDs to access the OneDep system. This authentication method enables contact authors to access sessions without sharing passwords. Using ORCiD with OneDep returns a summary table of all entries in which the ORCiD has been provided for the contact author.
wwPDB has improved the summary table with:
- Two new columns display the experimental method and the hold expiration date
- EMDB and BMRB accession codes for EM and NMR displayed alongside the entry ID
- All columns now sortable
While using a deposition ID and password to login into OneDep is still possible, we encourage depositors to use ORCiD for an improved user experience.
Questions or feedback? Contact [email protected].
12/12/2023
PDB Entries with Novel Ligands Now Distributed Only in PDBx/mmCIF and PDBML File Formats
The PDB three-character Chemical Component IDs are consumed and PDB has begun issuing five-character alphanumeric accession codes for CCD IDs in the OneDep system. To avoid confusion with current four-character PDB IDs, four-character codes are not used. Owing to limitations of the legacy PDB file format, PDB entries containing the new five character ID codes are distributed in PDBx/mmCIF and PDBML formats (see previous announcement).PDB entries containing these extended IDs will not be supported by the legacy PDB file format (see previous announcement).
In addition, wwPDB has reserved a set of CCD IDs: 01 - 99, DRG, INH, LIG that will never be used in the PDB. These reserved codes can be used for new ligands during structure determination so that they can be identified as new upon deposition and added to the CCD during biocuration.
wwPDB is asking users and software developers to review their code and remove any current limitations on PDB and CCD ID lengths, and to enable use of PDBx/mmCIF format files. https://github.com/wwPDB/extended-wwPDB-identifier-examplesExample files with extended PDB and/or CCD IDs are available via GitHub to assist with code revisions To learn about PDBx/mmCIF, please visit https://mmcif.wwpdb.org/.
For any further information please contact us at [email protected].
11/22/2023
Deprecation of FTP File Download Protocol in the PDB Archive
The FTP protocol for file downloads has been losing popularity over the years in favor of HTTP/S. There are many advantages of HTTP/S including speed, statelessness, security (HTTPS), and better support. Importantly during the past 2-3 years the main web browsers (Chrome and Firefox) have dropped support for the FTP protocol, which has effectively discontinued the FTP protocol for non-technical users.
Given that the majority of file download activity on the internet has moved to HTTP/S, wwPDB plans to deprecate FTP download protocol on November 1st 2024 (see previous announcement).
Support for the RSYNC protocol, which offers additional functionality, will continue to be maintained.
As announced previously, wwPDB supports protocol-specific DNS names:
Please contact [email protected] with any questions.
11/14/2023
Backbone Annotation and Standardization of Peptide Residues is Now Live
The wwPDB has rolled out updated Chemical Component Dictionary (CCD) data files with standardized atom naming and additional annotation of protein backbone and terminal atoms within peptide residues. Entries containing those updated CCDs have been updated accordingly. This improves the Findability and Interoperability of the PDB data and opens up new opportunities to use the updated peptide residue annotation.
As announced previously, new data items have been added to the CCD files for peptide-linking components to label atoms that form the backbone, N- or C-terminal groups. Three new CCD data items have been added to the CCD category _chem_comp_atom as pdbx_backbone_flag, pdbx_N-terminal_flag and pdbx_C-terminal_flag, flagging the backbone, N-terminal and C-terminal atoms, respectively.
Furthermore, we have standardized the atom nomenclature of peptide backbone atoms in CCD files to follow a standard convention. This follows a set of rules, outlined in our documentation, ensuring that atom nomenclature for carboxyl groups, amino groups and side chain linked carbons (C-alpha) follow a standard atom nomenclature. This allows clear identification of backbone atoms for peptide residues across the whole archive.
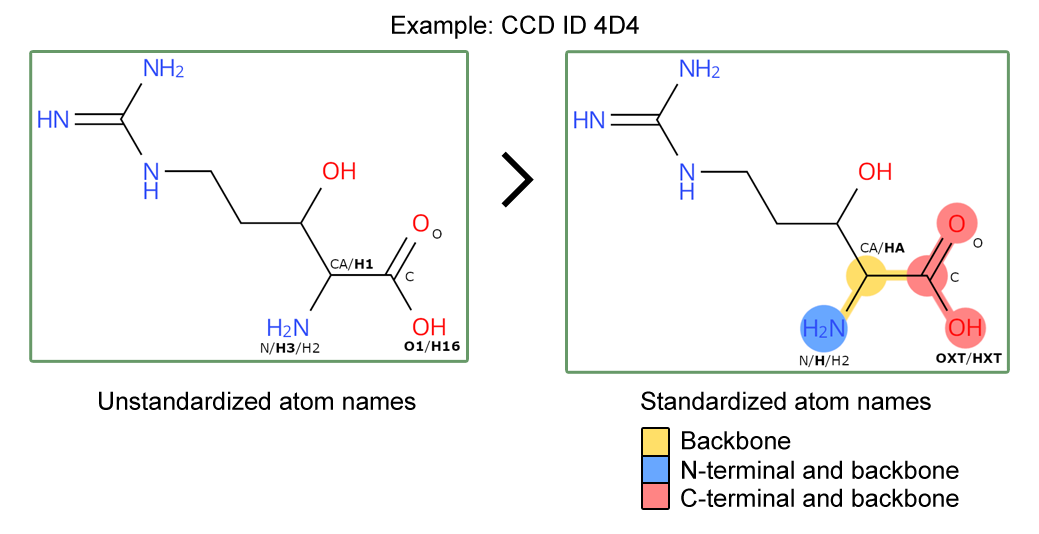 Updated peptide CCDs will have standardized atom names and backbone / N- and C- terminal annotation.
Updated peptide CCDs will have standardized atom names and backbone / N- and C- terminal annotation.Questions or feedback? Contact [email protected].
The peptide residues chemical component dictionary remediation project is part of the protein chemical modifications (PCMs) and post translational modifications (PTMs) remediation project, a wwPDB collaborative project carried out principally by PDBe at EMBL-EBI, and is funded by BBSRC grant number BB/V018779/1.
10/18/2023
Poster Prize Awarded at The Protein Society
The wwPDB Foundation made awards to outstanding student posters at the 2023 Annual Symposium of The Protein Society (July 13-16, Boston, MA).
 Taylor M. Laflamme
Taylor M. Laflamme Kevin Ramirez
Kevin RamirezTaylor M. Laflamme for
Specificity studies of small molecule inhibitors targeting the ankyrin repeat oncoprotein gankyrin
Taylor M. Laflamme (1), Emma I. Kane (1), Dipti Kanabar (2), Tejashri Chavan (2), Aaron Muth (2), & Donald E. Spratt (1)
1) Clark University; 2) St. John's University
Kevin Ramirez for
Host protein decoy fluorescence sensors for the detection of SARS-CoV-2 virions
Kevin Ramirez (1,2), David Bouzada (3), Arjan Bains (1,2), Mourad Sadqi (1,2), Eugenio Sentís-Vazquez(3), Patricia LiWang (1,2), and Victor Muñoz (1,2)
1) Department of Bioengineering, 2) CREST Center for Cellular & Biomolecular Machines, University of California Merced, 3) Universidad de Santiago de Compostela, Spain
Many thanks to The Protein Society organizers and poster prize judges for making these awards possible.
The wwPDB Foundation was established in 2010 to raise funds in support of the outreach activities of the wwPDB. The Foundation raised funds to help support PDB50 events, workshops, and educational publications. The Foundation is chartered as a 501(c)(3) entity exclusively for scientific, literary, charitable, and educational purposes.
The wwPDB Foundation is grateful for our industrial sponsors: Discngine, OpenEye Scientific, Roivant Sciences, Rigaku, and ThermoFisher Scientific. Individual sponsorships are also available.
Consider supporting the next 50 years of PDB's spirit of openness, cooperation, and education with a donation to the wwPDB Foundation.
10/11/2023
Poster Prize Awarded at IUCr
The wwPDB Foundation made awards to outstanding student posters at the 26th General Assembly and Congress of the International Union of Crystallography (IUCr) in Melbourne, Australia from August 22-29.
 Akila Pilapitiya
Akila Pilapitiya Liliana Guerrero
Liliana GuerreroAkila Pilapitiya for
The crystal structure of the toxin EspC from enteropathogenic Escherichia coli reveals approaches to combat diarrheal infections
Akila Pilapitiya, Lilian Hor, Jason Paxman, Begoña Heras
La Trobe University
Liliana Guerrero for
Drugging the Undruggable: Unveiling the Conformational Landscape and Ligandability of Phosphatases through Structural Biology
Liliana Guerrero (1), Ali Ebrahim (1), Blake T. Riley (1), Minyoung Kim (1,2), Qingqiu Huang (3), Aaron D. Finke (3), Daniel A. Keedy (1,4)
1) Structural Biology Initiative, CUNY Advanced Science Research Center 2) Department of Molecular Biology, Princeton University 3) Cornell High Energy Synchrotron Source (CHESS), Cornell University 4) Department of Chemistry and Biochemistry, City College of New York
Many thanks to the IUCr organizers and poster prize judges for making these awards possible.
The wwPDB Foundation was established in 2010 to raise funds in support of the outreach activities of the wwPDB. The Foundation raised funds to help support PDB50 events, workshops, and educational publications. The Foundation is chartered as a 501(c)(3) entity exclusively for scientific, literary, charitable, and educational purposes.
The wwPDB Foundation is grateful for our industrial sponsors: Discngine, OpenEye Scientific, Roivant Sciences, Rigaku, and ThermoFisher Scientific. Individual sponsorships are also available.
Consider supporting the next 50 years of PDB's spirit of openness, cooperation, and education with a donation to the wwPDB Foundation.
10/11/2023
Remediation of crystal structures deposited in non-standard coordinate frames
A total of 268 structure entries deposited to PDB in a non-standard coordinate frame or space group setting have been re-versioned and re-released between December 2022 and September 2023. The updates have been made to enable improved model visualization and validation against deposited experimental data using modern graphics and refinement software.
Each re-versioned entry has atom x,y,z coordinates transformed into the standard crystallographic frame. Non-crystal symmetry matrices, if present, have also been transformed to operate on the updated coordinates.
Coordinate transformations were extracted either from REMARK or SCALE records, or from transformations published in the primary citation.
All transformed structure coordinates have been carefully checked for integrity of crystal packing and where available, validation against the deposited structure factor data.
Original deposited coordinates/matrices remain accessible/downloadable as the previous major version in PDB's versioned archive.
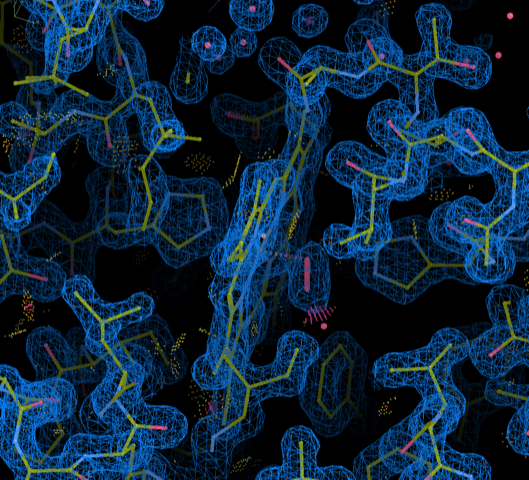 Coordinate frame-transformed myoglobin 1MBO coordinates with 2Fo-Fc map visualized using COOT.
Coordinate frame-transformed myoglobin 1MBO coordinates with 2Fo-Fc map visualized using COOT.
10/03/2023
Distribution of Electron Microscopy metadata in PDBx/mmCIF file format
At the beginning of October 2023, the Electron Microscopy Data Bank (EMDB) will start distributing electron microscopy (EM) metadata for its entries in PDBx/mmCIF file format alongside the already available XML formatted files. The addition of PDBx/mmCIF files in the archive will bring parity between EMDB and PDB (EM experimental data vs 3D structures) in the way metadata is served while improving the consistency and interoperability between the two archives. EMDB entries released before October 2023 will have PDBx/mmCIF files added at a later date. The description of PDBx/mmCIF categories and items featured in EM metadata files can be found in the PDBx/mmCIF dictionary (https://mmcif.wwpdb.org/dictionaries/mmcif_pdbx_v50.dic/Groups/em_group.html). For an EMDB entry EMD-xxxxx, PDBx/mmCIF file name will be emd-xxxxx.cif.gz and it will be located in the entry's subfolder called metadata (EMD-xxxxx/metadata) in the wwPDB archive repositories:
10/01/2023
Announcing Kyle Morris as EMDB's New Team Leader
wwPDB is excited to announce that Kyle Morris has joined the EMDB team as their new team leader. Kyle brings 10 years of experience in the cryo-EM field, and will draw on first-hand knowledge from his scientific research and staff roles within cryo-EM facilities. How the EMDB continues to engage with the community will be complemented with the experience Kyle gained developing and coordinating training programmes during his time at eBIC. His vision for EMDB includes strengthening its core functions, developing connections to complementary databases, and preparing the archive for future cryo-EM technologies, including machine learning applications.
09/12/2023
Coming Soon: PDB Entries with Novel Ligands Distributed Only in PDBx/mmCIF and PDBML File Formats
At current growth rates, we anticipate running out of three-character Chemical Component IDs by the end of 2023. After this point, the wwPDB will issue five-character alphanumeric accession codes for CCD IDs in the OneDep system. To avoid confusion with current four-character PDB IDs, four-character codes will not be used. Owing to limitations of the legacy PDB file format, PDB entries containing the new five character ID codes will only be distributed in PDBx/mmCIF and PDBML formats (see previous announcement).
In addition, wwPDB has reserved a set of CCD IDs: 01 - 99, DRG, INH, LIG that will never be used in the PDB. These reserved codes can be used for new ligands during structure determination so that they can be identified as new upon deposition and added to the CCD during biocuration.
wwPDB asks users and software developers to review code to remove any current limitations on CCD ID lengths, and to enable use of PDBx/mmCIF format files. Example files with extended CCD IDs are available via GitHub to assist code revisions. Information about the PDBx/mmCIF dictionary and file format is provided at mmcif.wwpdb.org.
For any further information please contact us at [email protected].
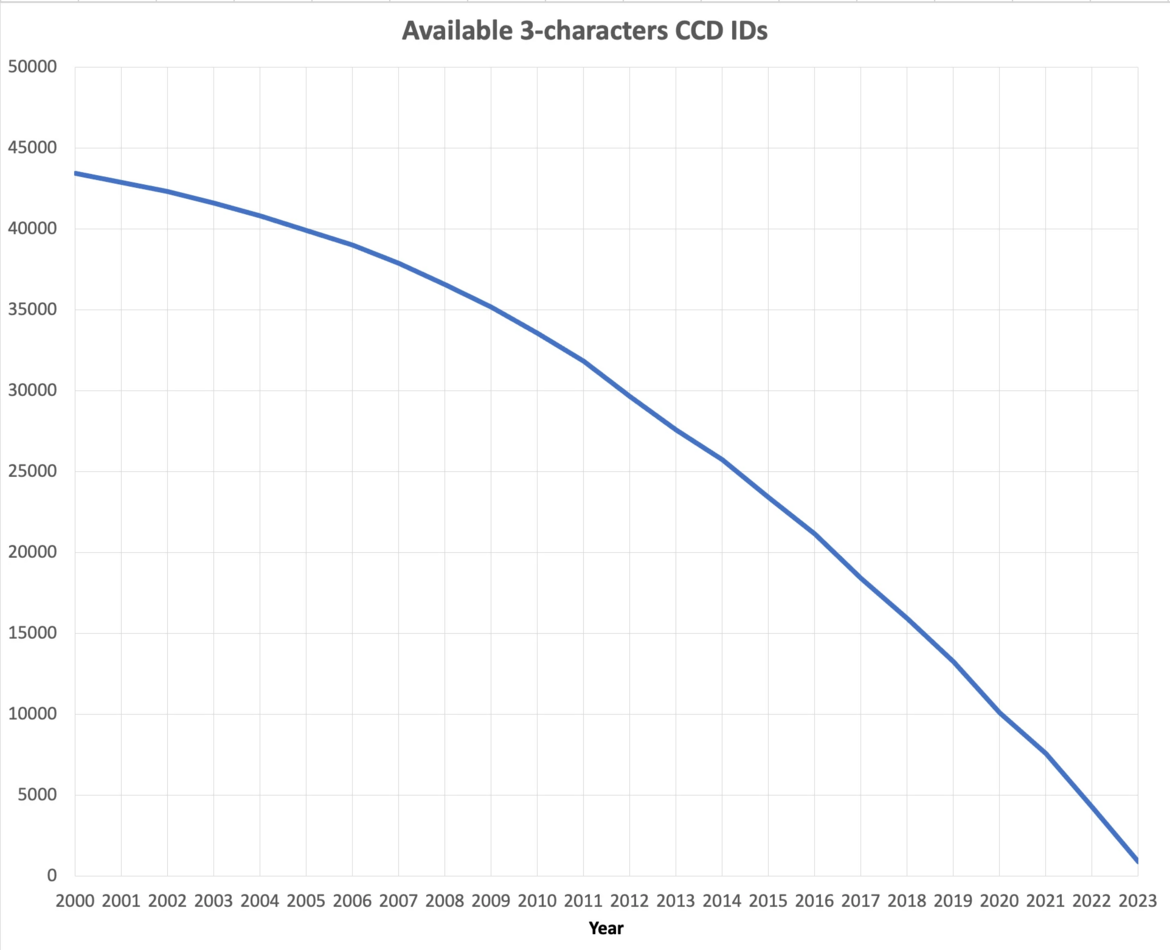 The number of available 3-character CCD IDs as of September 2023.
The number of available 3-character CCD IDs as of September 2023.
08/16/2023
wwPDB Events at IUCr
 wwPDB is celebrating its 20th anniversary.
wwPDB is celebrating its 20th anniversary.Meet wwPDB members from around the globe at the 26th General Assembly and Congress of the International Union of Crystallography (IUCr) in Melbourne, Australia from August 22-29.
Visit the wwPDB booth #26 in the exhibition hall and receive a special gift commemorating the 20th anniversary of the wwPDB. Learn about the latest wwPDB news, including Updated Annotation and Standardization of Peptide Residues, how to Access Depositions Using ORCiD, and more.
Other wwPDB events include:
- Wednesday August 23 in session A090: Biomolecular SAS and Integrative methods: Standards and Validation, Brinda Vallat (RCSB PDB) will speak about PDB-Dev: A prototype system for archiving integrative structures
- Saturday August 26: Director Stephen K. Burley will present his Keynote Lecture Protein Data Bank: From Two Pandemics to the Global Pandemic to mRNA Vaccines and Paxlovid
- Sunday August 27 in session A009/A010: Use and Comparison of Predicted Models from Primary Sequence in Structural Biology and Deep Learning & Artificial Intelligence in Structural Biology, Dennis Piehl (RCSB PDB) will describe Exploring experimentally-determined structures and computed structure models from artificial intelligence/machine learning at RCSB Protein Data Bank
- Tuesday August 29 in session A014: Databases and Data Management, Genji Kurisu (PDBj) will present Protein Data Bank Japan: the Asian hub of 3D macromolecular structural data and Christine Zardecki (RCSB PDB) will present RCSB Protein Data Bank: Sustaining a living digital data resource that enables breakthroughs in scientific research and biomedical education
The wwPDB Foundation will be sponsoring two awards of $250USD each that will be presented to the best posters from research presented in the Biological Macromolecule field by students (including undergraduates, postgraduates (Masters) and PhD) at the Congress.
Consider supporting 50 years of PDB's spirit of openness, cooperation, and education with a donation to the wwPDB Foundation. The wwPDB Foundation was established in 2010 to raise funds in support of the outreach activities of the wwPDB, including poster prizes that will be awarded throughout 2023.
 Visit the wwPDB booth to receive a wwPDB towel that celebrates the 20th anniversary of the partnership
Visit the wwPDB booth to receive a wwPDB towel that celebrates the 20th anniversary of the partnership
07/25/2023
Updated Annotation and Standardization of Peptide Residues
In October 2023, the wwPDB will roll out updated Chemical Component Dictionary (CCD) data files with standardized atom naming and additional annotation of protein backbone and terminal atoms within peptide residues. Entries containing those updated CCDs will also be updated accordingly. This will improve the Findability and Interoperability of the PDB data, as well as open up new opportunities to use the updated peptide residue annotation.
As part of this remediation process, we will add new data items to the CCD files for peptide-linking components to label atoms that form the backbone, N- or C-terminal groups. Three new CCD data items will be added to the CCD category _chem_comp_atom as pdbx_backbone_flag, pdbx_n-terminal_flag and pdbx_c-terminal_flag, flagging the backbone, N-terminal and C-terminal atoms, respectively.
Furthermore, we will be standardizing the atom nomenclature of peptide backbone atoms in CCD files to follow a standard convention. This will follow a set of rules, outlined in the documentation linked below, ensuring that atom nomenclature for carboxyl groups, amino groups and side chain linked carbons (C-alpha) follow a standard atom nomenclature. This will allow clear identification of backbone atoms for peptide residues across the whole archive.
Detailed information about this work is available from the wwPDB website, including PDBx/mmCIF dictionary extension and example files (GitHub; Peptide Residues Chemical Component Dictionary Remediation Documentation).
 Updated peptide CCDs will have standardized atom names and backbone / N- and C- terminal annotation.
Updated peptide CCDs will have standardized atom names and backbone / N- and C- terminal annotation.We encourage developers of software packages for refinement or visualization of PDB data to review this information.
Questions or feedback? Contact [email protected].
The peptide residues chemical component dictionary remediation project is part of the protein chemical modifications (PCMs) and post translational modifications (PTMs) remediation project, a wwPDB collaborative project carried out principally by PDBe at EMBL-EBI, and is funded by BBSRC grant number BB/V018779/1.
07/09/2023
Celebrating 20 Years of the wwPDB
 The Worldwide Protein Data Bank celebrates its 20th anniversary in 2023
The Worldwide Protein Data Bank celebrates its 20th anniversary in 2023In July 2003, the Worldwide Protein Data Bank was launched as a partnership between RCSB Protein Data Bank (RCSB PDB, USA), Protein Data Bank in Europe (PDBe), Protein Data Bank Japan (PDBj) in the management of the essential Protein Data Bank Core Archive of atomic-level, three-dimensional (3D) structures of biological macromolecule experimentally determined by macromolecular crystallography (MX), nuclear magnetic resonance (NMR) spectroscopy, or three-dimensional cryo-electron microscopy (3DEM; 1, 2).
Since then, the wwPDB collaboration has expanded to include additional Core Archives as partners: BioMagResBank (BMRB, USA) Core Archive of spectral and quantitative data derived from NMR spectroscopic investigations of biological macromolecules and metabolites; and Electron Microscopy Data Bank (EMDB, UK) Core Archives of 3D volumes and associated information of macromolecular complexes and subcellular structures from 3DEM and electron cryo tomography. wwPDB partners adhere to the FAIR principles of Findability, Accessibility, Interoperability, and Reusability (2), and ensure that all archival data can be accessed at no charge and with no limitations on usage under the most permissive Creative Commons CC0 1.0 Universal License.
As the wwPDB celebrates its twentieth year of operations, wwPDB is pleased to welcome Protein Data Bank China (PDBc) to the organization as an Associate Member. PDBc is based in the National Facility for Protein Science in Shanghai which is associated with Shanghai Advanced Research Institute of Chinese Academy of Sciences, and the SIAIS and iHuman Institutes of ShanghaiTech University. An announcement describing the process of PDBc joining the wwPDB has been submitted to Acta Cryst D (PDF, 3). PDBc has received training and support from the wwPDB partners, and it is expected that PDBc will process most, possibly all, depositions made by structural biologists working in the People’s Republic of China to all three wwPDB Core Archives.
Since its inception, the PDB has been an international archive and the establishment of the wwPDB ensured that valuable data contained in these Core Archives will continue to be stored, managed and kept freely available for the benefit of scientists worldwide.
- (1) Helen M. Berman, Kim Henrick, Haruki Nakamura. (2003) Announcing the worldwide Protein Data Bank. Nat Struct Biol 10: 980. doi: 10.1038/nsb1203-980
- (2) wwPDB consortium (2019) Protein Data Bank: the single global archive for 3D macromolecular structure data, Nucleic Acids Research 47:D520–D528 doi: 10.1093/nar/gky949
- (3) Wenqing Xu, Sameer Velankar, Ardan Patwardhan, Jeffrey C. Hoch, Stephen K. Burley, Genji Kurisu (2023) Announcing launch of Protein Data Bank China (PDBc) as an Associate Member of the Worldwide Protein Data Bank (wwPDB) Partnership (2023) Acta Cryst. D79 792-795 doi: Top
07/04/2023
PDB NextGen Archive Now Provides Intra-molecular Connectivity
Version 1.0 of the next generation archive repository (NextGen) for the PDB archive was made available in early 2023. This “NextGen” archive hosts enriched atomic coordinate files, in both PDBx/mmCIF and PDBML formats, with files available to download at files-nextgen.wwpdb.org.
The initial launch of the NextGen archive enriched coordinate files from the core PDB archive with sequence annotation from external resources such as UniProt, SCOP2 and Pfam at atom, residue, and chain levels. After consulting with user community, this release has added intra-molecular connectivity for each residue present in an entry, helping users transitioning from legacy PDB format to PDBx/mmCIF format. The connectivity information includes atom pairs, bond order, aromatic flag, and stereochemistry as incorporated from the PDB Chemical Component Dictionary (CCD). Users can extract this information from the _chem_comp_bond and _chem_comp_atom categories of the PDBx/mmCIF-formatted files from the NextGen archive.
To transition from legacy PDB format to PDBx/mmCIF, the file naming and data are structured based on extended PDB IDs with a two letter hash code, ‘third from last character' and 'second from last character’. This hash code will remain consistent once PDB ID codes are extended beyond four characters with the pdb_ prefix, e.g., PDB entry 8aly: https://files-nextgen.wwpdb.org/pdb_nextgen/data/entries/divided/al/pdb_00008aly/pdb_00008aly_xyz-enrich.cif.gz.
Users are encouraged to adopt PDBx/mmCIF format as early as possible. Learn more about PDBx/mmCIF format and related software resources at mmcif.wwpdb.org.
In the future, the PDB NextGen archive will continue to be updated with more enriched annotations from external database resources in the metadata, building on the content already provided in the structure model files in the PDB archive at files.wwpdb.org.
06/13/2023
wwPDB NMR restraint remediation conforms to standard NMR-STAR and NEF format
NMR data files are standardized in NEF and NMR-STAR format [Ulrich, 2019] for the PDB entries which have NMR data in many different software specific formats. This remediation project provides unified NMR data in single NMR-STAR/NEF file for the PDB entries. wwPDB is moving forward single file upload for NMR data (restraints, chemical shifts and possibly peak list) in NMR-STAR/NEF format and will eventually phase out the upload of software specific format in OneDep in future. During transition period, OneDep converts certain restraint formats into NMR-STAR/NEF at the deposition and provides users with NMR restraint validation in wwPDB validation reports for NMR entries having assigned chemical shifts and reasonable restraints in following supported formats:
AMBER, BIOSYM, CHARMM, CNS, CYANA, DYNAMO/TALOS/PALES, GROMACS, ISD, ROSETTA, SYBYL, and XPLOR-NIH
Depending on complexity of restraints and whether spectral peak lists have been deposited, the first rollout includes NMR entries with simple restraints in the supported formats and without spectral peak lists. The next rollout will include entries with spectral peak lists. Entries which met the following criteria are the scope for remediation:
- Both assigned chemical shifts and restraints have been deposited to the PDB
- A valid topology file or specific comments must be present to interpret restraint files in a specific format (i.e. AMBER, CHARMM, GROMACS)
- All atoms described in NMR data (assigned chemical shifts and restraints) are consistent with model’s atoms
- Sequence alignment between the model and restraints matches, allowing terminal sequence extensions
NMR-STAR format is the master format used to handle wide variety of restraints in this NMR restraint remediation project. Data items starting with “Auth_asym_ID”, “Auth_seq_ID”, “Auth_comp_ID”, and “Auth_atom_ID” terms, i.e. “_Gen_dist_constraint.Auth_seq_ID_1”, point to equivalent data items in mmCIF, “_atom_site.auth_asym_id”, “_atom_site.auth_seq_id”, “_atom_site.auth_comp_id”, “_atom_site.auth_atom_id”, respectively. Complete atom name mapping information is incorporated using “_Assembly” and “_Entity” categories. NMR constraints used by structure determination software but not covered by the NMR-STAR ontology, such as chemical planarity, equilibrium bond angle, non-crystallographic symmetry, etc., are stored as JSON data under "_Other_constraint_list.text_data" tag to ensure no information is lost during remediation. Furthermore, statistics on remediated restraints are available. NEF data files are provided as best effort conversions from the master NMR-STAR data files.
New unified NMR data files in NEF and NMR-STAR formats of existing NMR entries are distributed in the “nmr_data” directory of the PDB archive in the same manner for entries accompanied by the single NMR data file (https://files.wwpdb.org/pub/pdb/data/structures/divided/nmr_data/) and at BMRB archive (https://bmrb.io/ftp/pub/bmrb/nmr_pdb_integrated_data/coordinates_restraints_chemshifts/remediated_restraints). The existing data files in the PDB archive directories “nmr_chemical_shifts” and “nmr_restraints” are remained as is.
To retrieve NMR data, users should primarily obtain unified NMR data files at ../nmr_data directory. Conventional chemical shifts and restraints can be used only if unified NMR data are not available.
For newly deposited NMR entries with conventional separated NMR data files, OneDep now unifies assigned chemical shifts and restraints into a single data file, which can be accessed from OneDep upload summary page. After submission, depositors can not roll back to the conventional formats.
wwPDB validation reports are re-calculated to provide NMR restraint validation for the target entries. NMR restraints can be interpreted and weighted differently by various software during structure calculation. The NMR restraints validation software employs a more general approach recommended by NMR-VTF, independently verifying the satisfaction of each restraint in every model. This can occasionally result in a large number of violations when the actual structure determination software adopts a different approach or assigns less weight during structure calculation.
 Left chart: Distance Violation statistics for each model
Left chart: Distance Violation statistics for each model
Right chart: Distribution of distance restraints and violations in their respective categoriesFor any further information about NMR data remediation, please contact us at [email protected].
05/31/2023
DNS name changes for PDB archive downloads from wwPDB to start September 2023
In 2022, wwPDB introduced DNS names for programmatic access to PDB archive downloads:
- FTP: ftp://ftp.wwpdb.org
- HTTPS: https://files.wwpdb.org (replaces https://ftp.wwpdb.org)
- RSYNC: rsync://rsync.wwpdb.org (replaces rsync://wwpdb.org)
The PDB Archive Downloads documentation has detailed information.
Starting September 2023, wwPDB will start enforcing use of these updated DNS names. URLs in which the DNS name doesn’t match the protocol (e.g., https://ftp.wwpdb.org, ftp://files.wwpdb.org) will no longer work at that time.
Users who download PDB archive data programmatically are encouraged to switch to the new DNS names as soon as possible. HTTPS protocol is preferred (over FTP) for individual file downloads.
Please contact [email protected] with any questions.
05/16/2023
ls-lR index file to be removed July 12, 2023
With continuing growth of the PDB archive, the size of the file that lists all directory contents (currently https://files.wwpdb.org/pub/pdb/ls-lR) will become a challenge for long term maintenance. At 00:00 UTC on July 12, 2023, wwPDB will remove the following files from the PDB archive:
- ../pdb/ls-lR
- ../pdb/data/structures/ls-lR
- ../pdb/data/structures/models/ls-lR
- ../pdb/data/structures/models/current/ls-lR
- ../pdb/data/structures/models/obsolete/ls-lR
We strongly encourage users to utilize files previously announced that containing the same data (https://files.wwpdb.org/pub/pdb/holdings/).
These inventory data files offer a quick overview of data in the archive. Two new inventory files for experimental data are added. These files are in the extensible JSON format, and can be found under the new /pdb/holdings/ archive tree.
The inventory lists provided include:
- current_file_holdings.json.gz: a list of released PDB entries and the file types present for each in the PDB Core Archive (e.g. coordinate data, experimental data, validation report).
- refdata_id_list.json.gz: a list of released chemical reference entries, their content types (e.g., Chemical Component, BIRD), and the most recent modification date of the reference file.
- released_structures_last_modified_dates.json.gz: a list of released PDB entries with the most recent modification date of the PDBx/mmCIF file.
- released_experimental_data_last_modified_dates.json.gz: a list of released experimental data files with the most recent modification date
- obsolete_structures_last_modified_dates.json.gz: a list of obsoleted PDB entries with the most recent modification date of the PDBx/mmCIF file.
- obsolete_experimental_data_last_modified_dates.json.gz: a list of obsoleted experimental data files with the most recent modification date.
- all_removed_entries.json.gz: a list of obsoleted PDB entries including information for entry authors, entry title, release date, obsolete date, and superseding PDB ID, if any.
- unreleased_entries.json.gz: a list of on-hold PDB entries, their entry status, deposition date, and pre-release sequence information, where available.
Users are encouraged to utilize these inventory files. For example, checking for the update of the PDB archive can be performed using current_file_holdings.json.gz or released_structures_last_modified_dates.json.gz in /pub/pdb/holdings/.
Please contact [email protected] with any questions.
Updated May 30, 2023
05/05/2023
Prepare Depositions Using New pdb_extract Features
pdb_extract merges coordinate data, author-provided metadata, and data processing information from output files produced by structure determination programs into a complete PDBx/mmCIF file that can used for easy deposition with OneDep. Use the pdb_extract online form or the easily-installed command line interface that been re-engineered (Python).
Coordinate Data
Uploaded coordinate files (PDBx/mmCIF or PDB) will be checked against the PDBx/mmCIF dictionary. Legacy PDB formatted files will be converted to a OneDep-compliant PDBx/mmCIF data file.
Metadata
Depositors are encouraged to use the PDBj CIF editor to easily edit a template file to include corresponding metadata (sequence, crystallization condition, etc.). Method-specific templates have been pre-loaded into the PDBj CIF editor: X-ray, 3DEM, and NMR. Click on the top-left menu (light gray widget icon) to save the edited metadata file in PDBx/mmCIF. Upload this completed file in pdb_extract to prepare single or multiple related structures for submission.
Structure Determination Output Files
Upload the log file produced during data processing, and pdb_extract will parse the related diffraction metadata. Log files from various standalone packages and from CCP4 and autoPROC pipelines are supported, including:
- Aimless
- DIALS
- d*TREK
- HKL-2000
- HKL-3000
- Pointless
- Scala
- Scalepack
- XDS
- Xia2
- Xscale
04/24/2023
Poster Prize Awarded at #DiscoverBMB
The wwPDB Foundation made an award to for the best poster in the category Proteins: Structure, Function and Biophysics in the undergraduate competition at the #DiscoverBMB meeting hosted by the American Society for Biochemistry and Molecular Biology (ASBMB).
 Michael Quinteros and wwPDB Foundation Chair Celia Schiffer (University of Massachusetts Medical School)
Michael Quinteros and wwPDB Foundation Chair Celia Schiffer (University of Massachusetts Medical School)Michael Quinteros (Wesleyan University) presented “The mitochondrial Cu+ transporter PiC2 (SLC25A3) is a target of MTF1 and contributes to the development of skeletal muscle in vitro.”
This research was also published in “The mitochondrial Cu+ transporter PiC2 (SLC25A3) is a target of MTF1 and contributes to the development of skeletal muscle in vitro” by McCann C, Quinteros M, Adelugba I, Morgada MN, Castelblanco AR, Davis EJ, Lanzirotti A, Hainer SJ, Vila AJ, Navea JG, Padilla-Benavides T. (2022) Front Mol Biosci. 9:1037941 doi: 10.3389/fmolb.2022.1037941.
The wwPDB Foundation was established in 2010 to raise funds in support of the outreach activities of the wwPDB. The Foundation raised funds to help support PDB50 events, workshops, and educational publications. The Foundation is chartered as a 501(c)(3) entity exclusively for scientific, literary, charitable, and educational purposes.
The wwPDB Foundation is grateful for our industrial sponsors: Discngine, OpenEye Scientific, Roivant Sciences, Rigaku, and ThermoFisher Scientific. Individual sponsorships are also available.
Consider supporting the next 50 years of PDB's spirit of openness, cooperation, and education with a donation to the wwPDB Foundation.
04/02/2023
Removal of ls-lR index file from the PDB archive
With continuing growth of the PDB archive, the size of the file that lists all directory contents (currently https://files.wwpdb.org/pub/pdb/ls-lR) will become a challenge for long term maintenance. wwPDB plans to remove this file from the PDB archive at 00:00 UTC on July 12, 2023. We strongly encourage users to utilize files previously announced that containing the same data (https://files.wwpdb.org/pub/pdb/holdings/).
These inventory data files offer a quick overview of data in the archive. These files are in the extensible JSON format, and can be found under the new /pdb/holdings/ archive tree.
The inventory lists provided include:
- all_removed_entries.json.gz: a list of obsoleted PDB entries including information for entry authors, entry title, release date, obsolete date, and superseding PDB ID, if any.
- current_file_holdings.json.gz: a list of released PDB entries and the file types present for each in the PDB Core Archive (e.g. coordinate data, experimental data, validation report).
- obsolete_structures_last_modified_dates.json.gz: a list of obsoleted PDB entries with information about the most recent modification date of the PDBx/mmCIF file.
- refdata_id_list.json.gz: a list of released chemical reference entries, their content types (e.g., Chemical Component, BIRD), and the most recent modification date of the reference file.
- released_structures_last_modified_dates.json.gz: a list of released PDB entries with the most recent modification date of the PDBx/mmCIF file.
- unreleased_entries.json.gz: a list of on-hold PDB entries, their entry status, deposition date, and pre-release sequence information, where available.
Users are encouraged to utilize these inventory files. For example, checking for the update of the PDB archive can be performed using current_file_holdings.json.gz or released_structures_last_modified_dates.json.gz in /pub/pdb/holdings/.
Please contact [email protected] with any questions.
03/26/2023
Access Depositions Using ORCiD
We are pleased to announce that contact authors can now use ORCiDs to authenticate OneDep access. This authentication method allows each contact author to login to OneDep without the need for password sharing to view and access all their depositions.
OneDep login using a deposition ID and password is still possible, but will only provide access to the specific deposition.
Using ORCiD with OneDep returns a summary table of the entries in which the ORCiD has been provided for the contact author. Users can further access each of their entries’ deposition interfaces without the need to login again using a deposition ID or password.
 The ORCiD sign-in button is located below the existing login fields.
The ORCiD sign-in button is located below the existing login fields. After using the ORCiD login, this OneDep panel will display all available depositions.
After using the ORCiD login, this OneDep panel will display all available depositions.First-time OneDep contact authors will need to verify their email address before being able to create new depositions, similar to creating a new deposition without being logged-in with ORCiD.
Please be aware that adding a contact author ORCiD in the “Admin > Contact information” OneDep page will grant this author access to the current deposition.
Providing ORCiDs for OneDep contact authors has been mandatory since 2018.
03/09/2023
Tribute to Dr. Olga Kennard
The wwPDB consortium would like to pay tribute to Dr. Olga Kennard OBE FRS upon the sad news of her passing. Her pioneering work on the development of crystallographic databases laid the groundwork for modern molecular structure data archiving and the subsequent scientific breakthroughs that have made use of these data.
Olga was renowned for establishing the CCDC (Cambridge Crystallographic Data Centre) to maintain the Cambridge Structural Database (CSD) for small molecules. The CSD was first established by Olga in 1965, based on activities in her research group and has become the world’s repository for small-molecule organic and metal-organic crystal structures. Olga collected these data so that she could study how crystals form and her surveys were fundamental in the development of “crystal engineering”. Now containing over one million structures from X-ray and neutron diffraction analyses, this database of accurate 3D structures has become an essential resource to scientists around the world.
The increased interest and breakthroughs in solving biological molecular structures lead to the founding of the PDB (Protein Data Bank) by Walter Hamilton at BNL (Brookhaven National Laboratory). Olga worked with Walter to support the foundation of the PDB archive, with the archive initially operated jointly between BNL and CCDC (see the 1971 PDB announcement in Nature New Biology). While data processing was carried out at BNL, CCDC was responsible for organization of the data archive, with Olga and CCDC’s experience in data archiving hugely beneficial. Nowadays, the small molecules contained in biological structures archived in the PDB are validated using CCDC software which incorporates the knowledge embedded in the CSD.
 Left to right: Helen M. Berman, Janet Thornton, Shoshana Wodak, and Olga Kennard at the PDB-SwissProt Symposium in Jerusalem in 1996.
Left to right: Helen M. Berman, Janet Thornton, Shoshana Wodak, and Olga Kennard at the PDB-SwissProt Symposium in Jerusalem in 1996.Olga was a person of great integrity and drive and, in an age before computers had really developed, she saw the value of cross-data analysis to derive principles governing how small molecules interact. Very few scientists can claim that their work has enabled thousands of papers and investigations. Olga’s foresight and determination to establish and maintain the CSD means she is among those giants on whose shoulders many other scientists stand.
See also Celebrating Dr Olga Kennard OBE FRS, Founder of the Cambridge Structural Database, 1924 – 2023 at CCDC
03/07/2023
PDB entries with extended CCD or PDB IDs will be distributed in PDBx/mmCIF format only
wwPDB, in collaboration with the PDBx/mmCIF Working Group, has set plans to extend the length of accession codes (IDs) for PDB and Chemical Component Dictionary (CCD) entries in the future. PDB entries containing these extended IDs will not be supported by the legacy PDB file format. (see previous announcement)
CCD ID extension
CCD entries are currently identified by unique three-character alphanumeric IDs. At current growth rates, we anticipate running out of three-character IDs before 2024. After this point, the wwPDB will issue five-character alphanumeric accession codes for CCD IDs in the OneDep system. To avoid confusion with current four-character PDB IDs, four-character codes will not be used. Owing to limitations of the legacy PDB file format, PDB entries containing the new five character ID codes will only be distributed in PDBx/mmCIF format.
In addition, wwPDB has reserved a set of CCD IDs: 01 - 99, DRG, INH, LIG that will never be used in the PDB. These reserved codes can be used for new ligands during structure determination so that they can be identified as new upon deposition and added to the CCD during biocuration.
PDB ID extension
wwPDB will be extending PDB ID length to eight characters prefixed by ‘pdb’, e.g., pdb_00001abc. Each PDB entry has a corresponding Digital Object Identifier (DOI), often required for manuscript submission to journals and described in publications by the structure authors. Extended PDB IDs and corresponding PDB DOIs have been included in the PDBx/mmCIF formatted atomic coordinate files for all new and re-released entries since August 2021.
For example, PDB entry issued with 4-character PDB ID, 1abc, will have the extended PDB ID (pdb_00001abc) and corresponding PDB DOI (10.2210/pdb1abc/pdb), as listed in the _database_2 PDBx/mmCIF category.
loop_
_database_2.database_id
_database_2.database_code
_database_2.pdbx_database_accession
_database_2.pdbx_DOI
PDB 1abc pdb_00001abc 10.2210/pdb1abc/pdb
For example, PDB entry issued with 8-character PDB ID, pdb_00099xyz, after all 4-character IDs are consumed:
loop_
_database_2.database_id
_database_2.database_code
_database_2.pdbx_database_accession
_database_2.pdbx_DOI
PDB pdb_00099xyz pdb_00099xyz 10.2210/pdb_00099xyz/pdb
After all four-character PDB IDs are consumed, newly-deposited PDB entries will only be issued extended PDB ID codes, and PDB entries will only be distributed in PDBx/mmCIF format. PDB entries with four-character PDB IDs will remain unchanged.
Resources
wwPDB is asking users and software developers to review their code and remove any current limitations on PDB and CCD ID lengths, and to enable use of PDBx/mmCIF format files. Example files with extended PDB and/or CCD IDs are available via github to assist code revisions, see https://github.com/wwPDB/extended-wwPDB-identifier-examples. To learn about PDBx/mmCIF, please visit https://mmcif.wwpdb.org/.
For any further information please contact us at [email protected].
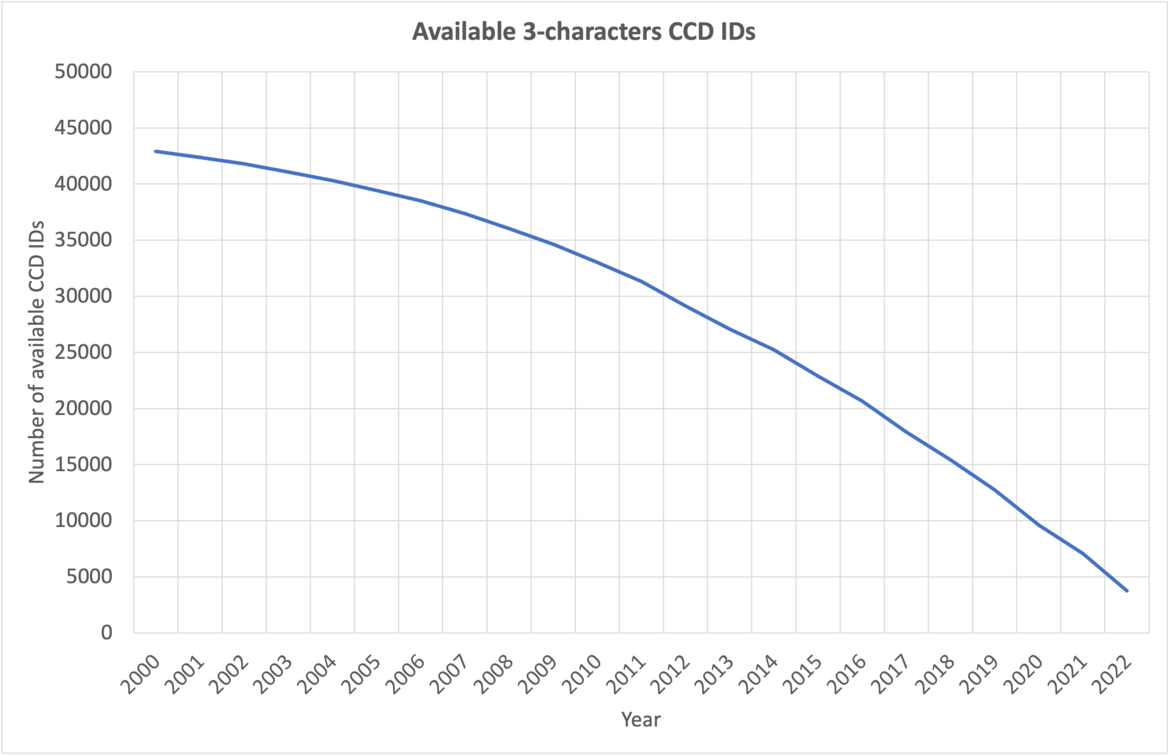 The number of available 3-character CCD IDs annually.
The number of available 3-character CCD IDs annually.
02/14/2023
Small Angle Scattering News
An outcome of a project aimed to test and benchmark different approaches for modeling SAS profiles from PDB coordinates has been published:
A round-robin approach provides a detailed assessment of biomolecular small-angle scattering data reproducibility and yields consensus curves for benchmarking
Trewhella, J., Vachette, P., Bierma, J., Blanchet, C., Brookes, E., Chakravarthy, S., Chatzimagas, L., Cleveland, T. E., Cowieson, N., Crossett, B., Duff, A. P., Franke, D., Gabel, F., Gillilan, R. E., Graewert, M., Grishaev, A., Guss, J. M., Hammel, M., Hopkins, J., Huang, Q., Hub, J. S., Hura, G. L., Irving, T. C., Jeffries, C. M., Jeong, C., Kirby, N., Krueger, S., Martel, A., Matsui, T., Li, N., Perez, J., Porcar, L., Prange, T., Rajkovic, I., Rocco, M., Rosenberg, D. J., Ryan, T. M., Seifert, S., Sekiguchi, H., Svergun, D., Teixeira, S., Thureau, A., Weiss, T. M., Whitten, A. E., Wood, K. & Zuo, X.
(2022) Acta Cryst. D78: 1315-1336 doi: 10.1107/S2059798322009184
In total, 171 SAXS and 76 SANS measurements for five proteins (ribonuclease A, lysozyme, xylanase, urate oxidase and xylose isomerase) were collected and analyzed centrally. In the process, new methods for data comparing and merging were developed. The data produced for this effort has been deposited in the SAS Biological Data Bank (SASBDB) as consensus data along with the contributing individual data sets.
In addition, a chapter describing the work done to establish the 2017 publication guidelines for biomolecular SAS, the establishment of the SASBDB, and the evolution and outcomes of the benchmarking project has been published:
Chapter One - Data quality assurance, model validation, and data sharing for biomolecular structures from small-angle scattering
Jill Trewhella
(2023) Methods in Enzymology 678: 1-22 doi: 10.1016/bs.mie.2022.11.002
These publications reflect the activities of the wwPDB Small Angle Scattering task force (SAStf) that first met with Chair Jill Trewhella in 2012. The SAStf was instrumental in progressing the important work that has led to biomolecular SAS being increasingly accepted as a mainstream structural biology technique.
02/06/2023
Prototype of PDB NextGen Archive now available
A prototype of a next generation archive repository for the PDB is now available. The archive, called “NextGen”, hosts structural model files in PDBx/mmCIF and PDBML formats at files-nextgen.wwpdb.org. This enriched PDB archive provides annotation from external database resources in the metadata in addition to the content provided in the structure model files in the PDB main archive at files.wwpdb.org.
This prototype provides sequence annotation from external resources such as UniProt, SCOP2 and Pfam at atom, residue, and chain levels. This mapping information is derived from the Structure Integration with Function, Taxonomy and Sequence (SIFTS) project (https://www.ebi.ac.uk/pdbe/docs/sifts/), a service developed and maintained by the PDBe and UniProt teams at EMBL-EBI. Sequence mappings are provided in _pdbx_sifts_unp_segments and _pdbx_sifts_xref_db_segments categories for each segment, _pdbx_sifts_xref_db at residue level, and _atom_site at atom level.
The PDB NextGen Repository is currently updated monthly on the first Wednesday of the month at 00:00 UTC and is subject to change in the future. You can access these NextGen files at the following locations:
Data are structured based on entry ID with a two letter hash code, ‘third from last character' and 'second from last character’. This hash code will remain consistent once PDB ID codes are extended beyond four characters with the pdb_ prefix.
Some examples are shown below:
Access entry pdb_00008aly at https://files-nextgen.wwpdb.org/pdb_nextgen/data/entries/divided/al/pdb_00008aly/Both PDBx/mmCIF and PDBML are provided at this location. For entry pdb_00008aly:- pdb_00008aly_xyz-enrich.cif.gz
-
pdb_00008aly_xyz-no-atom-enrich.xml.gz
Please contact [email protected] with any questions.
01/31/2023
Enhanced Collection of Starting Models
A new PDBx/mmCIF category, _pdbx_initial_refinement_model has been introduced to improve information collected about starting model for X-ray, 3DEM and NMR methods.
Experimentally derived vs computed models will be distinguished. Provenances of the resources where the starting model was obtained (e.g., PDB, AlphaFoldDB, RoseTTAFold, etc.) and its accession code/identifier will be captured, if publicly available.
For the full definition, see pdbx_initial_refinement_model. An example is below:
_pdbx_initial_refinement_model.id 1
_pdbx_initial_refinement_model.entity_id_list 1
_pdbx_initial_refinement_model.type 'experimental model'
_pdbx_initial_refinement_model.source_name PDB
_pdbx_initial_refinement_model.accession_code 3LTQ
wwPDB strongly recommends all PDB users and software developers to review their code and adopt this definition for future applications.
01/30/2023
Structure Predictors: Use ModelCIF for Computed Structure Models
ModelCIF (GitHub) is a data information framework developed for and by computational structural biologists to describe structural models of macromolecules derived from computational methods. It provides an extensible data representation for deposition, archiving, and public dissemination of these models of proteins to enable delivery of Findable, Accessible, Interoperable, and Reusable (FAIR) data to users worldwide.
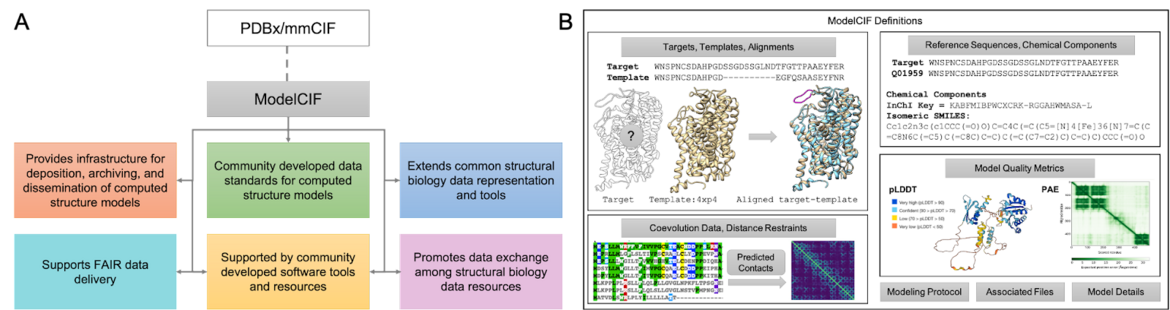 A. Overview of the ModelCIF extension of PDBx/mmCIF. B. Schematic representation of ModelCIF data specifications. ModelCIF includes definitions for input data used in template-based and template-free modeling; reference information for macromolecular sequences and small molecule components; local and global CSM quality metrics; and metadata information regarding modeling protocol, CSM classification (ab initio, homology, etc.) and descriptions of associated files.
A. Overview of the ModelCIF extension of PDBx/mmCIF. B. Schematic representation of ModelCIF data specifications. ModelCIF includes definitions for input data used in template-based and template-free modeling; reference information for macromolecular sequences and small molecule components; local and global CSM quality metrics; and metadata information regarding modeling protocol, CSM classification (ab initio, homology, etc.) and descriptions of associated files.ModelCIF is an extension of the Protein Data Bank Exchange/macromolecular Crystallographic Information Framework (PDBx/mmCIF), which is the global data standard for representing experimentally-determined, three-dimensional (3D) structures of macromolecules and associated metadata. The PDBx/mmCIF framework and its extensions (e.g., ModelCIF) are managed by the wwPDB in collaboration with relevant community stakeholders such as the wwPDB ModelCIF Working Group.
This semantically rich and extensible data framework for representing computed structure models (CSMs) accelerates the pace of scientific discovery. Furthermore, use of this data standard promotes interoperation among structural biology data resources, with ModelCIF currently used by the ModelArchive, AlphaFold DB, and MODBASE repositories. A manuscript was recently submitted to bioRxiv describing the architecture, contents, and governance of ModelCIF as well as tools and processes for maintaining and extending the data standard [1].
Visit the ModelCIF GitHub for more information about this data information framework.
[1} Vallat B, Tauriello G, Bienert S, Haas J, Webb BM, et al. ModelCIF: An extension of PDBx/mmCIF data representation for computed structure models. bioRxiv doi: 10.1101/2022.12.06.518550.
01/10/2023
PDB Reaches a New Milestone: 200,000+ Entries
 Depositors: Download this image, write the number of structures deposited, and tag us in your photos
Depositors: Download this image, write the number of structures deposited, and tag us in your photosWith this week's update, the PDB archive contains a record 200,069 entries. The archive passed 150,000 structures in 2019 and 100,000 structures in 2014.
Established in 1971, this central, public archive has reached this critical milestone thanks to the efforts of structural biologists throughout the world who contribute their experimentally-determined protein and nucleic acid structure data.
wwPDB data centers support online access to three-dimensional structures of biological macromolecules that help researchers understand many facets of biomedicine, agriculture, and ecology, from protein synthesis to health and disease to biological energy. Many milestones have been reached since the archive released the 100,000th structure in 2014. PDB data have been seminal in understanding SARS-CoV-2, and provided the foundation for the development of AI/ML techniques for predicting protein structure. The 50th anniversary of the PDB was celebrated throughout 2021.
Today, the archive is quite large, containing more than 3,000,000 files related to these PDB entries that require more than 1086 Gbytes of storage. PDB structures contain more than 1.8 billion non-hydrogen atoms.
Function follows form
In the 1950s, scientists had their first direct look at the structures of proteins and DNA at the atomic level. Determination of these early three-dimensional structures by X-ray crystallography ushered in a new era in biology-one driven by the intimate link between form and biological function. As the value of archiving and sharing these data were quickly recognized by the scientific community, the Protein Data Bank (PDB) was established as the first open access digital resource in all of biology by an international collaboration in 1971 with data centers located in the US and the UK.
Among the first structures deposited in the PDB were those of myoglobin and hemoglobin, two oxygen-binding molecules whose structures were elucidated by Chemistry Nobel Laureates John Kendrew and Max Perutz. With this week's regular update, the PDB welcomes 266 new structures into the archive. These structures join others vital to drug discovery, bioinformatics and education.
The PDB is growing rapidly, increasing in size by ~160% since 2011 (doubling in size every 6-8 years). In 2022, an average of 275 new structures were released to the scientific community each week. The resource is accessed hundreds of millions of times annually by researchers, students, and educators intent on exploring how different proteins are related to one another, to clarify fundamental biological mechanisms and discover new medicines.
Twenty Years of Collaboration
Since its inception, the PDB has been a community-driven enterprise, evolving into a mission critical international resource for biological research. The wwPDB partnership was established in July 2003 with PDBe, PDBj, and RCSB PDB. Today, the collaboration includes partners BMRB (joined in 2006) and EMDB (2021).
The wwPDB ensures that these valuable PDB data are securely stored, expertly managed, and made freely available for the benefit of scientists and educators around the globe. wwPDB data centers work closely with community experts to define deposition and annotation policies, resolve data representation issues, and implement community validation standards. In addition, the wwPDB works to raise the profile of structural biology with increasingly broad audiences.
Each structure submitted to the archive is carefully curated by wwPDB staff before release. New depositions are checked and enhanced with value-added annotations and linked with other important biological data to ensure that PDB structures are discoverable and interpretable by users with a wide range of backgrounds and interests.
wwPDB eagerly awaits the next 100,000 structures and the invaluable knowledge these new data will bring.
01/03/2023
Time-stamped Copies of PDB and EMDB Archives
 A snapshot of the PDB Core Archive as of January 2, 2023 is available.
A snapshot of the PDB Core Archive as of January 2, 2023 is available. A snapshot of the PDB Core archive (ftp://ftp.wwpdb.org, https://s3.rcsb.org) as of January 2, 2023 has been added to ftp://snapshots.wwpdb.org, https://s3snapshots.rcsb.org (AWS), and ftp://snapshots.pdbj.org. Snapshots have been archived annually since 2005 to provide readily identifiable data sets for research on the PDB archive.
The directory 20230102 includes the 199,755 experimentally-determined structure and experimental data available at that time. Atomic coordinate and related metadata are available in PDBx/mmCIF, PDB, and XML file formats. The date and time stamp of each file indicates the last time the file was modified. The snapshot of PDB Core Archive is 1086 GB.
A snapshot of the EMDB Core archive (ftp://ftp.ebi.ac.uk/pub/databases/emdb/) as of January 2, 2023 can be found in ftp://ftp.ebi.ac.uk/pub/databases/emdb_vault/20230102/ and ftp://snapshots.pdbj.org/20230102/. The snapshot of EMDB Core Archive contains map files and their metadata within XML files for both released and obsoleted entries (24186 and 262, respectively) and is 8.9 TB in size.