wwPDB 2022 News
Contents
12/15/2022
PDB Certified as a Global Core Biodata Resource
The Global Biodata Coalition (GBC) today announced that the Protein Data Bank (PDB) archive is included in the first list of Global Core Biodata Resources (GCBRs)—a collection of 37 resources whose long term funding and sustainability is critical to life science and biomedical research worldwide.
The PDB is one of three core archives jointly managed by the Worldwide Protein Data Bank (wwPDB) partnership. Three wwPDB members, RCSB Protein Data Bank (RCSB PDB), Protein Data Bank in Europe (PDBe) and Protein Data Bank Japan (PDBj) share deposition, biocuration and validation responsibilities contributing to the management of this single global archive of 3D biostructure data for macromolecules. RCSB PDB also serves as the wwPDB-designated PDB Archive Keeper.
Researchers the world over rely on data resources to professionally manage, curate and freely disseminate research data. These resources include deposition databases which archive and preserve primary research data, and knowledge bases which collate and add value to the archived data through expert curation and annotation, enabling those data to be mined, combined and used to advance research.
Like keystone species in an ecosystem, GCBRs represent the most crucial components or nodes within the global life science data infrastructure, whose failure would have a critically adverse impact on global research endeavors. A key property of the GCBRs is that the data they hold is available openly and can be accessed and used without restriction by researchers the world over, a principle adhered to by the PDB archive throughout its over 50 year history.
The GBC brings together major public and charitable funders, who are committed to work in partnership to develop more sustainable funding approaches to support biodata resources—many of which currently rely on short-term, unstable funding.
This first collection of GCBRs were selected through a rigorous two-stage process open to biodata resources globally (see Global Core Biodata Resources: Concept and Selection Process). More than 60 resources submitted expressions of interest, for which it was necessary that they met several eligibility criteria. At each stage of the selection process, the candidate biodata resources were assessed by a panel of more than 50 independent expert reviewers against a series of criteria that included their scientific focus, the size and reach of their user communities, their quality of service, their governance, and their impact on global research.
 Visit the Global Biodata Coalition for details.
Visit the Global Biodata Coalition for details.
11/01/2022
Deprecation of FTP File Download Protocol in the PDB Archive
The FTP protocol for file downloads has been losing popularity over the years in favor of HTTP/S. There are many advantages of HTTP/S including speed, statelessness, security (HTTPS), and better support. Importantly during the past 2-3 years the main web browsers (Chrome and Firefox) have dropped support for the FTP protocol, which has effectively discontinued the FTP protocol for non-technical users.
Given that the majority of file download activity on the internet has moved to HTTP/S, wwPDB plans to deprecate FTP download protocol on November 1st 2024.
wwPDB has traditionally supported FTP, together with HTTP/S and RSYNC. Gradual deprecation of the FTP protocol, in favor of the HTTP/HTTPS protocol will be approached while maintaining support for the RSYNC protocol which offers additional functionality compared to the other 2 protocols.
As announced previously, we have introduced DNS names that are specific to the protocols:
- files.wwpdb.org for HTTP/S
- ftp.wwpdb.org for FTP. To be deprecated on November 1st 2024. Note that from September 2023 this DNS name will not accept HTTP/S traffic.
- rsync.wwpdb.org for RSYNC
Starting September 2023, wwPDB will start enforcing use of these updated DNS names for the preparation of FTP protocol deprecation.
Please contact [email protected] with any questions.
10/12/2022
wwPDB Charter: Full and Associate Members
Founded in 2003, the Worldwide Protein Data Bank (wwPDB) is dedicated to archiving, management, and public dissemination of structural biology data, and is committed to the FAIR Principles (Findability, Accessibility, Interoperability, Reusability) that are emblematic of responsible stewardship of public domain information.
wwPDB operations are governed by the wwPDB Charter (PDF), which was most recently renewed in 2021. The renewed agreement provides for two types of wwPDB membership--Full and Associate.
Current wwPDB Full Members include three founding members [Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB, USA), Protein Data Bank in Europe (PDBe, United Kingdom), and Protein Data Bank Japan (PDBj, Japan)] and two specialist data resource members [Biological Magnetic Resonance Bank (BMRB, USA and Japan; joined in 2006) and Electron Microscopy Data Bank (EMDB, United Kingdom; joined in 2021)].
Full Members jointly manage three wwPDB Core Archives, including the Protein Data Bank, the Biological Magnetic Resonance Bank, and the Electron Microscopy Data Bank. Data safety and security and periodic archival updates are the primary responsibility of three wwPDB-designated Archive Keepers, including RCSB PDB for the Protein Data Bank, BMRB for the Biological Magnetic Resonance Bank, and EMDB for the Electron Microscopy Data Bank.
The wwPDB acknowledges the importance of global equity in the ability to deposit and access data, and the need for international involvement and collaboration in maintaining the wwPDB Core Archives.
The renewed wwPDB Charter describes the Associate Membership program, wherein Associate Members are expected to contribute to some of the wwPDB activities (deposition, validation, biocuration, remediation, storage, dissemination of public-domain structural biology data stored in one or more wwPDB Core Archives). The responsibilities of an individual wwPDB Associate Member for deposition, validation, biocuration, remediation, storage, and/or dissemination of public-domain structural biology data stored in one or more wwPDB Core Archives must be agreed upon by all wwPDB Full Members.
At the discretion of wwPDB Full Members, an external organization may be invited to apply to become a wwPDB Associate Member, following preliminary discussions and successful completion of due diligence and demonstration of sufficient technical expertise, adequate infrastructure, and sustainable funding. The decision to admit a new wwPDB Associate Member must be by unanimous vote of the current Heads of the existing wwPDB Full Members, supported by a simple majority of the voting members of the wwPDB Advisory Committee.
Full details of the arrangement are provided in the wwPDB Charter (PDF).
09/26/2022
Improved EM validation with Q-score
As announced previously, wwPDB validation of 3DEM structures for which there is both a model and an EM volume will include the Q-score metric (Pintilie, G., et al., 2020, Nat. Methods). This follows recommendations from the wwPDB/EMDB workshop on cryo-EM data management, deposition and validation in 2020 (white paper in preparation), as well as EM Validation Challenge events (Lawson C., et al., 2020, Struct. Dyn.; Lawson, et al., 2021, Nat. Methods). This will be the first quantitative parameter of residue and chain resolvability for EM maps in wwPDB validation reports and will provide an additional map-model assessment criterion.
The Q-score calculates the resolvability of atoms by measuring similarity of the map values around each atom relative to a Gaussian-like function for a well resolved atom. Q-score of 1 indicates that the similarity is perfect whilst closer to 0 indicates the similarity is low. If the atom is not well placed in the map then a negative Q-score value may be reported. Therefore, Q-score values in the reports will be in a range of -1 to +1.
The wwPDB EM validation reports will provide Q-scores for single particle, helical reconstruction, electron crystallography and subtomogram averaging entries for which both an EM map and coordinate model have been deposited.
Validation reports (PDF files) will contain images of the average per-residue Q-scores color-mapped onto ribbon models with views from three orthogonal directions. Similar images will also be introduced to visualize the per-residue atom-inclusion scores. Comparison of these two sets of images will assist in visual assessment of the model-to-map fit and quality.
The images below show the model with each residue colored according to its Q-score.
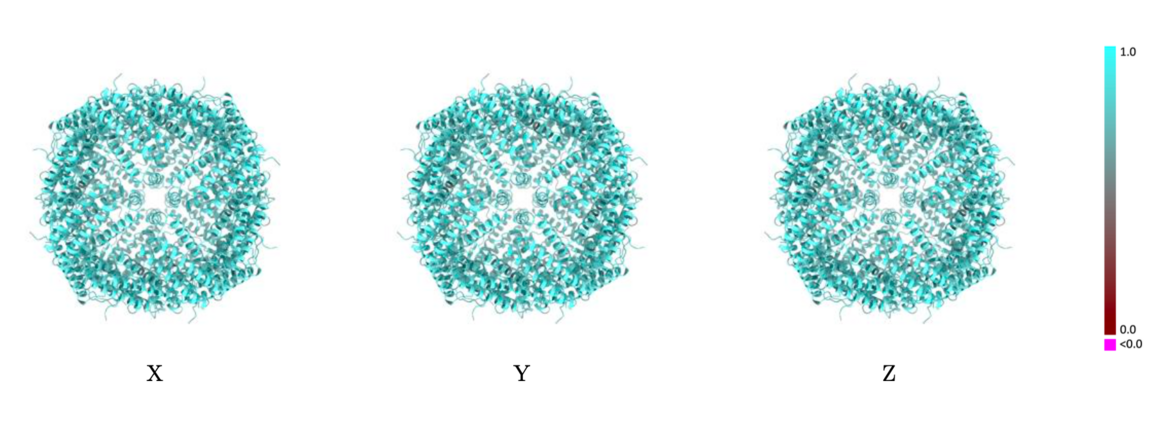 Example showing mostly cyan colors indicating Q-score closer to 1 and a good resolvability of atoms
Example showing mostly cyan colors indicating Q-score closer to 1 and a good resolvability of atoms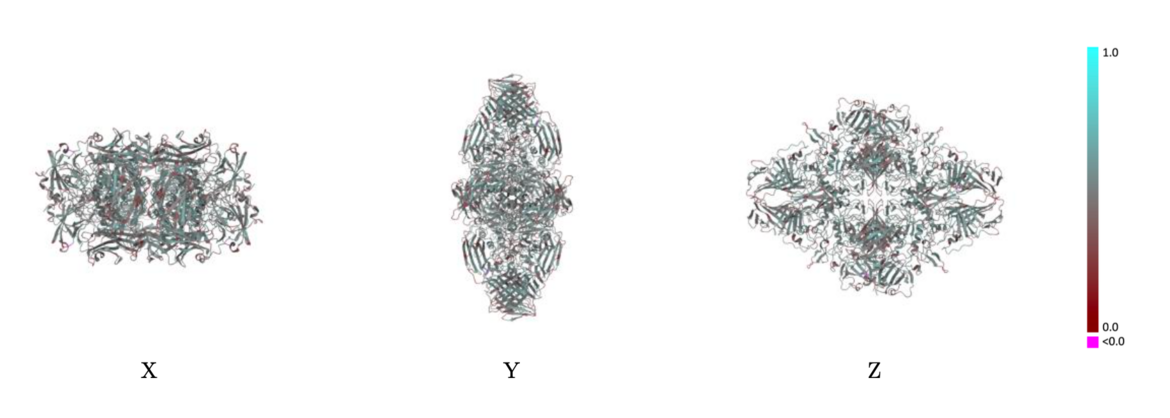 Example showing mostly red colors indicating Q-score closer to 0 and not a good resolvability of atoms
Example showing mostly red colors indicating Q-score closer to 0 and not a good resolvability of atomsThe validation reports will also contain a table of average per-chain values of both metrics (Q-score and atom inclusion) as well as their overall average values for the entire model.
The per-residue and the per-chain average atom-inclusion and Q-score values will also be provided in the mmCIF and XML formatted validation files. The mmCIF categories _pdbx_vrpt_summary_entity_fit_to_map and _pdbx_vrpt_model_instance_map_fitting will be introduced to include both the Q-scores and atom-inclusion values. The existing items, _pdbx_vrpt_summary_entity_geometry.average_residue_inclusion and _pdbx_vrpt_model_instance_geometry.residue_inclusion for atom inclusion will no longer be used.
The PDB Core Archive holds validation reports that assess each 3DEM model in the PDB along with the associated experimental 3D volume in EMDB. Validation reports of 3DEM structures (map and model) can be downloaded at the following wwPDB mirrors:
The EMDB Core Archive holds validation reports that assess each EMDB map/tomogram entry. Validation reports for all EMDB volumes can be downloaded at the following wwPDB mirrors:
Additional information about validation reports is available for EM map+model, EM map-only, and EM tomograms.
If you have any questions or queries about wwPDB validation, please contact us at [email protected].
09/05/2022
Future Planning: PDB entries with extended CCD or PDB IDs will be distributed in the PDBx/mmCIF format only
wwPDB, in collaboration with the PDBx/mmCIF Working Group, has set plans to extend the length of accession codes (IDs) for PDB and Chemical Component Dictionary (CCD) entries in the future. PDB entries containing these extended IDs will not be supported by the legacy PDB file format. (see previous announcement)
CCD ID extension
CCD entries are currently identified by unique three-character alphanumeric IDs. At current growth rates, we anticipate running out of three-character IDs before 2024. After this point, the wwPDB will issue five-character alphanumeric accession codes for CCD IDs in the OneDep system. To avoid confusion with current four-character PDB IDs, four-character codes will not be used. Owing to limitations of the legacy PDB file format, PDB entries containing the new five character ID codes will only be distributed in PDBx/mmCIF format.
In addition, wwPDB has reserved a set of CCD IDs: 01 - 99, DRG, INH, LIG that will never be used in the PDB. These reserved codes can be used for new ligands during structure determination so that they can be identified as new upon deposition and added to the CCD during biocuration.
PDB ID extension
wwPDB will be extending PDB ID length to eight characters prefixed by ‘pdb’, e.g., pdb_00001abc. Each PDB entry has a corresponding Digital Object Identifier (DOI), often required for manuscript submission to journals and described in publications by the structure authors. Extended PDB IDs and corresponding PDB DOIs have been included in the PDBx/mmCIF formatted atomic coordinate files for all new and re-released entries since August 2021.
For example, PDB entry issued with 4-character PDB ID, 1abc, will have the extended PDB ID (pdb_00001abc) and corresponding PDB DOI (10.2210/pdb1abc/pdb), as listed in the _database_2 PDBx/mmCIF category.
loop_
_database_2.database_id
_database_2.database_code
_database_2.pdbx_database_accession
_database_2.pdbx_DOI
PDB 1abc pdb_00001abc 10.2210/pdb1abc/pdb
For example, PDB entry issued with 8-character PDB ID, pdb_00099xyz, after all 4-character IDs are consumed:
loop_
_database_2.database_id
_database_2.database_code
_database_2.pdbx_database_accession
_database_2.pdbx_DOI
PDB pdb_00099xyz pdb_00099xyz 10.2210/pdb_00099xyz/pdb
After all four-character PDB IDs are consumed, newly-deposited PDB entries will only be issued extended PDB ID codes, and PDB entries will only be distributed in PDBx/mmCIF format. PDB entries with four-character PDB IDs will remain unchanged.
Resources
wwPDB is asking users and software developers to review their code and remove any current limitations on PDB and CCD ID lengths, and to enable use of PDBx/mmCIF format files. Example files with extended PDB and/or CCD IDs are available via github to assist code revisions, see https://github.com/wwPDB/extended-wwPDB-identifier-examples. To learn about PDBx/mmCIF, please visit https://mmcif.wwpdb.org/.
For any further information please contact us at [email protected].
 The number of available 3-character CCD IDs annually.
The number of available 3-character CCD IDs annually.
08/22/2022
DNS name changes for PDB archive downloads from wwPDB
wwPDB has introduced DNS names for programmatic access to PDB archive downloads:
- FTP: ftp://ftp.wwpdb.org
- HTTPS: https://files.wwpdb.org (replaces https://ftp.wwpdb.org)
- RSYNC: rsync://rsync.wwpdb.org (replaces rsync://wwpdb.org)
The PDB Archive Downloads documentation has detailed information.
Starting September 2023, wwPDB will start enforcing use of these updated DNS names. URLs in which the DNS name doesn’t match the protocol (e.g., https://ftp.wwpdb.org, ftp://files.wwpdb.org) will no longer work at that time.
Users who download PDB archive data programmatically are encouraged to switch to the new DNS names as soon as possible. HTTPS protocol is preferred (over FTP) for individual file downloads.
Please contact [email protected] with any questions.
07/25/2022
Improved EM validation with Q-score
Starting September 23, wwPDB validation of 3DEM structures for which there is both a model and an EM volume will include the Q-score metric (Pintilie, G., et al., 2020, Nat. Methods). This follows recommendations from the wwPDB/EMDB workshop on cryo-EM data management, deposition and validation in 2020 (white paper in preparation), as well as EM Validation Challenge events (Lawson C., et al., 2020, Struct. Dyn.; Lawson, et al., 2021, Nat. Methods). This will be the first quantitative parameter of residue and chain resolvability for EM maps in wwPDB validation reports and will provide an additional map-model assessment criterion.
The Q-score calculates the resolvability of atoms by measuring similarity of the map values around each atom relative to a Gaussian-like function for a well resolved atom. Q-score of 1 indicates that the similarity is perfect whilst closer to 0 indicates the similarity is low. If the atom is not well placed in the map then a negative Q-score value may be reported. Therefore, Q-score values in the reports will be in a range of -1 to +1.
The wwPDB EM validation reports will provide Q-scores for single particle, helical reconstruction, electron crystallography and subtomogram averaging entries for which both an EM map and coordinate model have been deposited.
Validation reports (PDF files) will contain images of the average per-residue Q-scores color-mapped onto ribbon models with views from three orthogonal directions. Similar images will also be introduced to visualize the per-residue atom-inclusion scores. Comparison of these two sets of images will assist in visual assessment of the model-to-map fit and quality.
The images below show the model with each residue colored according to its Q-score.
 Example showing mostly cyan colors indicating Q-score closer to 1 and a good resolvability of atoms
Example showing mostly cyan colors indicating Q-score closer to 1 and a good resolvability of atoms Example showing mostly red colors indicating Q-score closer to 0 and not a good resolvability of atoms
Example showing mostly red colors indicating Q-score closer to 0 and not a good resolvability of atomsThe validation reports will also contain a table of average per-chain values of both metrics (Q-score and atom inclusion) as well as their overall average values for the entire model.
The per-residue and the per-chain average atom-inclusion and Q-score values will also be provided in the mmCIF and XML formatted validation files. The mmCIF categories _pdbx_vrpt_summary_entity_fit_to_map and _pdbx_vrpt_model_instance_map_fitting will be introduced to include both the Q-scores and atom-inclusion values. The existing items, _pdbx_vrpt_summary_entity_geometry.average_residue_inclusion and _pdbx_vrpt_model_instance_geometry.residue_inclusion for atom inclusion will no longer be used.
The PDB Core Archive holds validation reports that assess each 3DEM model in the PDB along with the associated experimental 3D volume in EMDB. Validation reports of 3DEM structures (map and model) can be downloaded at the following wwPDB mirrors:
The EMDB Core Archive holds validation reports that assess each EMDB map/tomogram entry. Validation reports for all EMDB volumes can be downloaded at the following wwPDB mirrors:
Additional information about validation reports is available for EM map+model, EM map-only, and EM tomograms.
If you have any questions or queries about wwPDB validation, please contact us at [email protected].
06/28/2022
New Publication: PDBx/mmCIF Ecosystem
 An article on the PDBx/mmCIF Ecosystem is part of a special issue on "Computational Resources for Molecular Biology"
An article on the PDBx/mmCIF Ecosystem is part of a special issue on "Computational Resources for Molecular Biology"A new article by the wwPDB and the PDBx/mmCIF Working Group describes the community-driven data representation for structural biology data that is critical to the PDB archive. It describes file standards and governance, and summarizes software tools for data processing and checking.
PDBx/mmCIF Ecosystem: Foundational Semantic Tools for Structural Biology
John D. Westbrook, Jasmine Y. Young, Chenghua Shao, Zukang Feng, Vladimir Guranovic, Catherine L. Lawson, Brinda Vallat, Paul D. Adams, John M. Berrisford, Gerard Bricogne, Kay Diederichs, Robbie P. Joosten, Peter Keller, Nigel W. Moriarty, Oleg V. Sobolev, Sameer Velankar, Clemens Vonrhein, David G. Waterman, Genji Kurisu, Helen M. Berman, Stephen K. Burley, Ezra Peisach
(2022) Journal of Molecular Biology 434: 167599 doi: 10.1016/j.jmb.2022.167599
This article is dedicated to John D. Westbrook, whose work established the PDBx/mmCIF data dictionary and format as the foundation of the modern Protein Data Bank (PDB) archive (wwPDB.org).
05/03/2022
Distributing PDBx/mmCIF-Formatted Assembly Files
Starting May 3, 2022, the PDB archive distributes assembly files in PDBx/mmCIF format, allowing direct access and visualization of the curated assemblies for all PDB entries (original announcement).
Previously, PDBx/mmCIF formatted assembly files provided for structures were non-PDB compliant, however the coordinates use model numbers to differentiate alternate symmetry copies of PDB chain IDs. This method is not ideal, nor necessary, for the current archive PDBx/mmCIF format and has led to limited use of these files in community software tools. In response to this issue and recommendations by the wwPDB advisory committee, we are implementing updated, standardized practices for generation of assembly files for all PDB entries.
These updated PDBx/mmCIF format assembly files have improved organization of assembly data to support usage by the community. These files will include all symmetry generated copies of each chain within a single model, with distinct chain IDs (_atom_site.auth_asym_id and _atom_site.label_asym_id) assigned to each. Generation of distinct chain IDs in assembly files are based upon the following rules:
Chain IDs of the original chains from the atomic coordinate file will be retained (e.g., A) Assign unique chain ID (atom_site.label_asym_id and atom_site.auth_asym_id) for each symmetry copy within a single model. Rules of chain ID assignments: - The applied index of the symmetry operator (pdbx_struct_oper_list.id) will be appended to the original chain ID separated by a dash (e.g., A-2, A-3, etc.)
- If there are more than one type of symmetry operators applied to generate symmetry copy, a dash sign will be used between two operators (e.g., A-12-60, A-60-88, etc.)
In addition, entity ID and chain ID mapping categories are provided: _pdbx_entity_remapping and _pdbx_chain_remapping.
A new directory (ftp.wwpdb.org/pub/pdb/data/assemblies/mmCIF/) was created for the distribution of these updated assembly files. The directory containing the existing assembly mmCIF files for large entries has been removed (ftp.wwpdb.org/pub/pdb/data/biounit/mmCIF/'>ftp.wwpdb.org/pub/pdb/data/biounit/mmCIF/).
wwPDB asks all PDB users and software developers to review code and address any limitations related to PDB assemblies. Sample files were made available for testing purposes and to support community adoption at GitHub.com/wwpdb/assembly-mmcif-examples.
If you plan to use these assembly files for graphical viewing, check if your visualization software (e.g., PyMol, ChimeraX, etc.) supports instantiation of assemblies directly from atomic coordinate files (_struct_assembly related categories), for improved efficiency.
For any further information please email [email protected].
03/18/2022
ModelCIF Extension for Computed Structure Models
ModelCIF, an extension of PDBx/mmCIF for computed structure models, is now available. The PDBx/mmCIF data standard underpins the Protein Data Bank (PDB) Core Archive, which is jointly managed by the worldwide Protein Data Bank (wwPDB) consortium. A software library called python-modelcif has been developed to support ModelCIF and enables reading and writing mmCIF files compliant with ModelCIF.
ModelCIF serves as the data standard for representing structural models of macromolecules obtained using computational methods. These computed structure models may be derived from existing structure templates using homology or comparative modeling or can be obtained from ab initio modeling methods. ModelCIF data standard is being adopted by computational biologists as well as major repositories of computed structure models, including ModelArchive, MODBASE, and AlphaFoldDB Protein Structure Database repositories for computed structure models. Partial support for ModelCIF is also available in SWISS-MODEL projects and will soon be added to the SWISS-MODEL Repository.
ModelCIF is developed and maintained by the wwPDB ModelCIF Working Group (WG), consisting of representatives from the wwPDB and the computational structural biology community. The WG is focused on developing common data standards and software tools for archiving and visualization of computed structure models. The WG promotes adoption of ModelCIF within the computational modeling community, and is also involved in developing software tools that support ModelCIF. Research teams making computed structure models available from their own web portals are strongly encouraged to do so using the ModelCIF data standard and integrate them into the 3D-Beacons network. Structural biologists are strongly encouraged to deposit computed structure models to the ModelArchive to ensure long-term preservation and public access. Guidelines on how to deposit computed structure models together with relevant metadata are also available on ELIXIR's RDMkit page for structural bioinformatics.
 ModelCIF Working Group
ModelCIF Working Group
03/16/2022
CASP15 Call for Targets
CASP (Critical Assessment of protein Structure Prediction) is in search for targets for the upcoming CASP15 modeling experiment (starting in May 2022). CASP community experiments aim to advance the state of the art in protein structure modeling. Every other year since 1994, CASP collects information on soon-to-be released experimental structures, passes on sequence data to the structure modeling community, and collects blind predictions of structure for assessment. Typically, about 100 modeling groups from around the world participate. Results of CASP experiments are assessed by leaders in the field (Independent Assessors), and published in special issues of the journal PROTEINS.
Following the 2020 CASP14 experiment, it is hard to find a structural biologist who has not heard about the success of deep learning methods in modeling protein structures, particularly by the AlphaFold and more recently RosettaFold. As a result of these advances, computed protein structures are becoming much more widely used in a broadening range of applications. Since CASP14, the protein modeling community has intensified development of these methods and extended their application to include modeling of protein complexes and protein ensembles. CASP15 will provide definitive insight into how successful these new developments are.
CASP15’s success depends on generosity of the experimental community in providing targets as ground truth against which to assess the computation methods. Over the years more than 150 structure determination groups have provided over 1100 targets for CASP challenges. For CASP15, we are requesting submission of all types of experimental structures determined by X-ray crystallography, cryo-electron microscopy and NMR as potential targets, but are particularly interested in the following:
- High resolution structures of single proteins. Because of the high accuracy of the new computational methods, it is becoming difficult to distinguish experimental error from computational error in low resolution structures.
- Structures with few or no known sequence relatives. Consistently accurate computed structures for this class of target requires methods that do not depend on evolutionary relationships.
- Protein complexes. Deep learning methods already show increased performance in this area, and a range of complexes is needed to establish exactly how powerful these are. Assessment will be in partnership with CAPRI, as in other recent CASPs.
- RNA structures, RNA complexes, and protein RNA complexes. Many more RNA structures are now being determined experimentally, opening this area for more extensive rigorous assessment.
- Proteins with clearly determined alternative conformations. An obvious extension beyond single protein structures is the calculation of ensembles of conformations. The new computational methods are already being applied to this problem, but there is a paucity of definitive experimental data to assess these against, which may limit this category.
- Protein-organic ligand complexes. Deep learning methods are also being applied to these structures. We are exploring including this category in CASP15. A major challenge is obtaining suitable targets.
CASP also plans to include modeling assisted by sparse experimental data, in collaboration with experimental groups in NMR, SAXS, and crosslinking mass spectrometry. For that, protein material is needed (this is not expected for most targets, but if available, it would be much appreciated!).
So, if you have suitable targets in any if these areas, we would very much appreciate you getting in touch by replying to this email or writing to [email protected] or suggesting your target directly through the CASP15 target entry page.
Note that CASP target providers are regularly invited to contribute to CASP special journal issue papers (e.g. Computational models in the service of X-ray and cryo-electron microscopy structure determination (2021) Proteins 89: 1633-1646; Target highlights in CASP14: Analysis of models by structure providers (2021) Proteins 89: 1647-1672), and we plan to continue this practice in the future.
02/25/2022
Deposition of Half-maps for Certain EM Entries Now Mandatory
As announced previously, deposition of half-maps for single-particle, single-particle-based helical, and sub-tomogram averaging reconstructions to the EM Data Bank (EMDB) is mandatory as of February 25, 2022. This change is in response to a long-standing community request to the wwPDB EMDB Core Archive and was also a recommendation from the 2020 wwPDB single-particle cryo-EM data-management workshop (white paper in preparation). Several recommendations from this workshop have already been implemented in the wwPDB OneDep system. These include improvements to wwPDB validation reports and enhancements for capturing metadata via the deposition interface.
Mandatory half-maps must be unfiltered, unmasked, unsharpened, and positioned in the same coordinate-space and orientation as the primary map such that they superimpose. The availability of half-maps will contribute to improved validation of EM structures as reflected in the wwPDB validation reports.
wwPDB strongly urges developers of cryo-EM processing software for the affected modalities to implement support for output of such half-maps (if this is not already available).
Any queries about this policy change can be directed to [email protected].
02/01/2022
Distributing PDBx/mmCIF-Formatted Assembly Files
Starting May 3, 2022, the PDB archive will distribute assembly files in PDBx/mmCIF format, allowing direct access and visualization of the curated assemblies for all PDB entries.
Currently, PDBx/mmCIF formatted assembly files are provided for structures that are non-PDB compliant, however the coordinates use model numbers to differentiate alternate symmetry copies of PDB chain IDs. This method is not ideal, nor necessary, for the current archive PDBx/mmCIF format and has lead to limited use of these files in community software tools. In response to this issue and recommendations by the wwPDB advisory committee, we are implementing updated, standardized practices for generation of assembly files for all PDB entries.
These updated PDBx/mmCIF format assembly files will have improved organization of assembly data to support usage by the community. These files will include all symmetry generated copies of each chain within a single model, with distinct chain IDs (_atom_site.auth_asym_id and _atom_site.label_asym_id) assigned to each. Generation of distinct chain IDs in assembly files are based upon the following rules:
Chain IDs of the original chains from the atomic coordinate file will be retained (e.g., A) Assign unique chain ID (atom_site.label_asym_id and atom_site.auth_asym_id) for each symmetry copy within a single model. Rules of chain ID assignments: - The applied index of the symmetry operator (pdbx_struct_oper_list.id) will be appended to the original chain ID separated by a dash (e.g., A-2, A-3, etc.)
- If there are more than one type of symmetry operators applied to generate symmetry copy, a dash sign will be used between two operators (e.g., A-12-60, A-60-88, etc.)
In addition, entity ID and chain ID mapping categories will be provided: _pdbx_entity_remapping and _pdbx_chain_remapping.
A new directory (ftp.wwpdb.org/pub/pdb/data/assemblies/mmCIF/) will be created for the distribution of these updated assembly files. The directory containing the existing assembly mmCIF files for large entries will be removed (ftp.wwpdb.org/pub/pdb/data/biounit/mmCIF/).
wwPDB asks all PDB users and software developers to review code and address any limitations related to PDB assemblies. Sample files are made available for testing purposes and to support community adoption at GitHub.com/wwpdb/assembly-mmcif-examples.
If you plan to use these assembly files for graphical viewing, check if your visualization software (e.g., PyMol, ChimeraX, etc.) supports instantiation of assemblies directly from atomic coordinate files (_struct_assembly related categories), you do so for improved efficiency.
For any further information please email [email protected].
01/18/2022
Improved support for extended PDBx/mmCIF structure factor files
Extensions to the PDBx/mmCIF dictionary for reflection data with anisotropic diffraction limits, for unmerged reflection data, and for quality metrics of anomalous diffraction data are now supported in OneDep.
In October 2020, a subgroup of the wwPDB PDBx/mmCIF Working Group was convened to develop a richer description of experimental data and associated data quality metrics. Members of this Data Collection and Processing Subgroup are all actively engaged in development and support of diffraction data processing software. The Subgroup met virtually for several months discussing, reviewing, and finalizing a new set dictionary content extension that were incorporated into the PDBx/mmCIF dictionary on February 16, 2021. A reference implementation of the new content extensions has been developed by Global Phasing Ltd.
These extensions facilitate the deposition and archiving of a broader range of diffraction data, as well as new quality metrics pertaining to these data. These extensions cover three main areas:
- scaled and merged reflection data that have been processed to take account of diffraction anisotropy, by providing descriptors for that anisotropy, in terms of (1) a parameter-free definition of a cut-off surface by means of a per-reflection “signal” and a threshold value for that signal, and (2) the ellipsoid providing the best fit to the resulting cut-off surface;
- scaled and unmerged reflection data, by providing extra item definitions aimed at ensuring that such data can be meaningfully re-analysed, and their quality assessed independently from the associated model, after retrieval from the archive;
- anomalous diffraction data, by adding descriptors for numerous relevant, but previously missing, statistics.
The new mmCIF data extensions describing anisotropic diffraction now enable archiving of the results of Global Phasing’s STARANISO program. Developers of other software can make use of them or extend the present definitions to suit their applications. Example files created by autoPROC, BUSTER (version 20210224) and Gemmi that are compliant with the new dictionary extensions are provided in a GitHub repository.
These example files, and similarly compliant files produced by other data processing and/or refinement programs, are suitable for direct uploading to the wwPDB OneDep system. Automatic recognition of that compliance, implemented by means of explicit dictionary versioning using the new pdbx_audit_conform record, will avoid unnecessary pre-processing at the time of deposition. This improved OneDep support will ensure a lossless round trip between data processing/refinement in the lab and deposition at the PDB.
wwPDB strongly encourages structural biologists to always use the latest versions of structure determination software packages to produce data files for PDB deposition. wwPDB also encourages crystallographers wishing to deposit new structures together with their associated diffraction data to use the software which guarantees consistency between data and final model. This consistency is difficult to achieve when separate diffraction data files and model coordinate files are pieced together a posteriori by ad hoc means.
wwPDB also encourages depositors to make their raw diffraction images available from one of the public repositories to allow direct access to the original diffraction image data.
01/05/2022
Time-stamped Copies of PDB and EMDB Archives
 A snapshot of the PDB Core Archive (ftp://ftp.wwpdb.org) as of January 3rd, 2022 has been added to ftp://snapshots.wwpdb.org and ftp://snapshots.pdbj.org.
A snapshot of the PDB Core Archive (ftp://ftp.wwpdb.org) as of January 3rd, 2022 has been added to ftp://snapshots.wwpdb.org and ftp://snapshots.pdbj.org. A snapshot of the PDB Core archive (ftp://ftp.wwpdb.org) as of January 3, 2022 has been added to ftp://snapshots.wwpdb.org and ftp://snapshots.pdbj.org. Snapshots have been archived annually since 2005 to provide readily identifiable data sets for research on the PDB archive.
The directory 20220103 includes the 185541 experimentally-determined structure and experimental data available at that time. Atomic coordinate and related metadata are available in PDBx/mmCIF, PDB, and XML file formats. The date and time stamp of each file indicates the last time the file was modified. The snapshot of PDB Core Archive is 923 GB.
A snapshot of the EMDB Core archive (ftp://ftp.ebi.ac.uk/pub/databases/emdb/) as of January 3, 2022 can be found in ftp://ftp.ebi.ac.uk/pub/databases/emdb_vault/20220103/ and ftp://snapshots.pdbj.org/20220103/. The snapshot of EMDB Core Archive contains map files and their metadata within XML files for both released and obsoleted entries (18059 and 254, respectively) and is 4.5 TB in size.